
1 The Challenge
Effective therapeutics for prevalent diseases require deep insight into molecular, genetic, and cellular factors, yet this knowledge is scattered across diverse sources, posing major challenges for data integration and analysis.

2 Our Solution
Here, we present CROssBARv2, a heterogeneous knowledge graph (KG) based system to facilitate systems biology and drug discovery/repurposing. CROssBARv2 collects large-scale biological data from 32 data sources and stores them in a Neo4j-based graph database.

3 Content
CROssBARv2 consists of 2,709,502 nodes and 12,688,124 relationships between 14 node types (i.e., protein, gene, organism, domain, biological process, molecular function, cellular component, drug, compound, disease, pathway, phenotype, EC number, and side effect).

4 Easy Access
We developed a large language model interface to convert natural language queries into Neo4j's Cypher query language back and forth to access information within the KG and answer user questions without LLM hallucinations. Other means of interacting with CROssBSRv2 are our GraphQL API and the Neo4j browser.

CROssBARv2 is expected to contribute to life sciences research considering (i) the discovery of biological mechanisms at the molecular level and (ii) the development of effective therapeutic strategies.
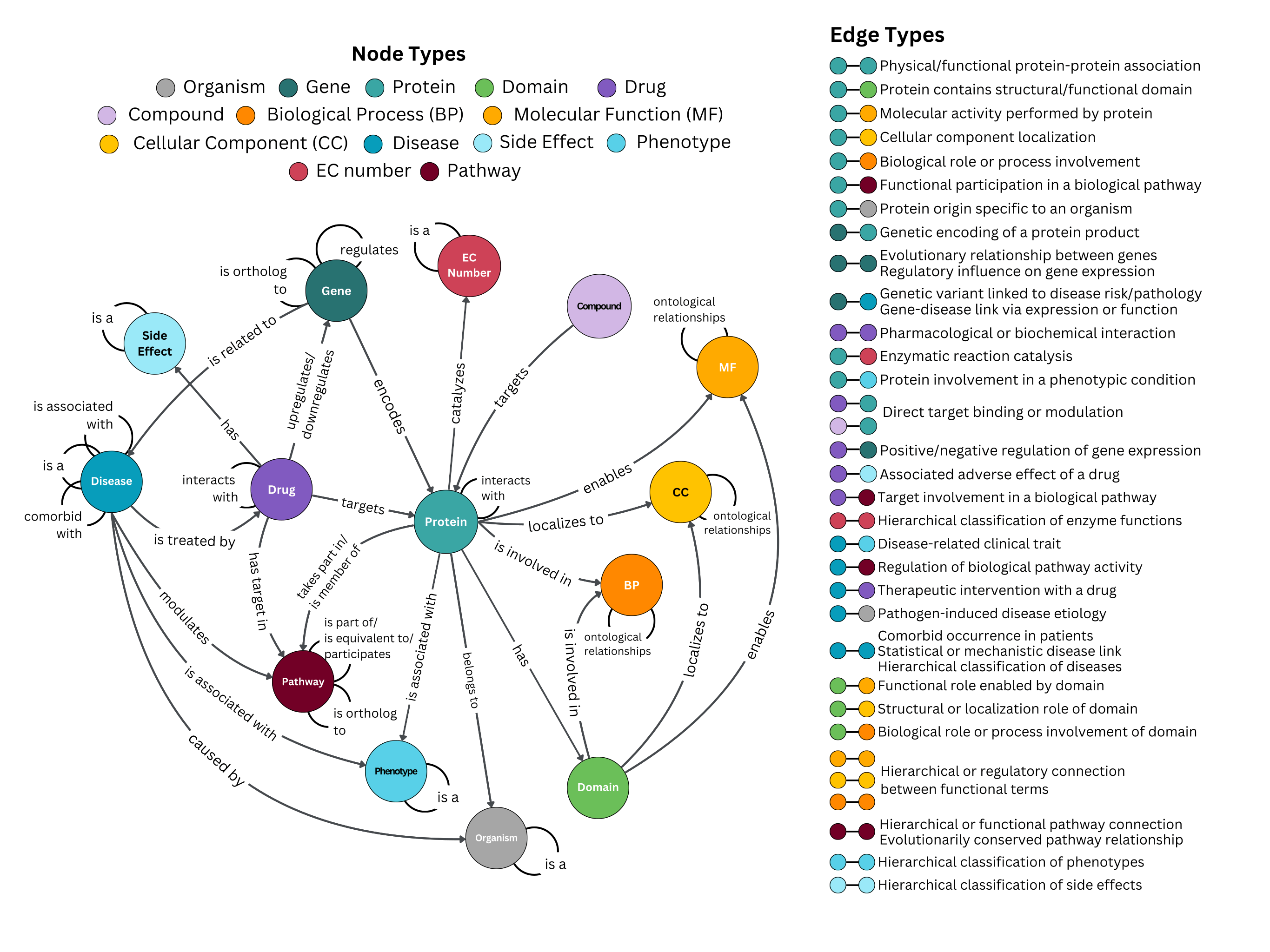
| Node | Identifier | Source Database | ||
| 1 | Protein | UniProt | UniProt | |
| 2 | Gene | Entrez | UniProt | |
| 3 | Organism Taxon | NCBI Tax ID | UniProt | |
| 4 | Drug | Drugbank | Drugbank | |
| 5 | Compound | ChEMBL | ChEMBL | |
| 6 | Side effect | Meddra | SIDER, OffSIDES, ADReCS | |
| 7 | Disease | MONDO | MONDO | |
| 8 | Phenotype | HPO | HPO | |
| 9 | Protein Domain | InterPro | InterPro | |
| 10 | Pathway | KEGG, Reactome | KEGG, Reactome | |
| 11 | EC number | EC number | Expasy ENZYME | |
| 12 | Molecular function | GO | GOA | |
| 13 | Cellular component | GO | GOA | |
| 14 | Biological process | GO | GOA |
| Source node type | Relationship | Target node type | Source node identifier | Target node identifier | Source database | Filtering (thresholds for filtering) | ||
| 1 | Protein | Interacts with | Protein | UniProt | UniProt | STRING, BioGRID, IntAct | Only high-confidence interactions (score >0.7) from STRING | |
| 2 | Protein | Has | Protein Domain | UniProt | InterPro | InterPro | - | |
| 3 | Protein | Enables / Contributes to | Molecular Function | UniProt | GO | GOA | electronic annotations (i.e., IEA) filtered out | |
| 4 | Protein | Located in / Is active in / Part of | Cellular component | UniProt | GO | GOA | electronic annotations (i.e., IEA) filtered out | |
| 5 | Protein | Involved in | Biological process | UniProt | GO | GOA | electronic annotations (i.e., IEA) filtered out | |
| 6 | Protein | Take part in | Pathway | UniProt | KEGG, Reactome | Reactome | electronic annotations (i.e., IEA) filtered out | |
| 7 | Protein | Catalyzes | EC Number | UniProt | EC Number | Expasy ENZYME | - | |
| 8 | Protein | Belongs to | Organism Taxon | UniProt | NCBI Tax ID | UniProt | - | |
| 9 | Protein | Is associated with | Phenotype | UniProt | HPO | HPO | - | |
| 10 | Gene | Encodes | Protein | Entrez | UniProt | UniProt | - | |
| 11 | Gene | Is ortholog to | Gene | Entrez | Entrez | OMA, Pharos | orthologs between human and model organisms | |
| 12 | Gene | Regulates | Gene | Entrez | Entrez | DoRothEA, TRRUST, CollecTRI | conflicting regulatory modes (i.e., activation, repression, and unknown or activation and repression) were excluded | |
| 13 | Gene | Is related to | Disease | Entrez | MONDO | ClinVar, DisGeNET, KEGG, DISEASES, humsavar, OpenTargets | - | |
| 14 | Drug | Interacts with | Drug | Drugbank | Drugbank | DDInter, KEGG | - | |
| 15 | Drug | Targets | Protein | Drugbank | UniProt | Drugbank, Pharos, ChEMBL, DGIdb, KEGG, STITCH | - | |
| 16 | Drug | Upregulates / Downregulates | Gene | Drugbank | Entrez | CTD | contradictory expression annotations (i.e., both increase and decrease expression) were excluded | |
| 17 | Drug | Has | Side effect | Drugbank | Meddra | SIDER, OffSIDES, ADReCS | - | |
| 18 | Drug | Has target in | Pathway | Drugbank | KEGG | KEGG, Reactome | - | |
| 19 | Compound | Targets | Protein | ChEMBL | UniProt | ChEMBL, STITCH | - | |
| 20 | Disease | Is comorbid with | Disease | MONDO | MONDO | Malacards | - | |
| 21 | Disease | Is associated with | Disease | MONDO | MONDO | DisGeNET | - | |
| 22 | Disease | Is a | Disease | MONDO | MONDO | MONDO | - | |
| 23 | Disease | Is associated with | Phenotype | MONDO | HPO | HPO | electronic annotations (i.e., IEA) filtered out | |
| 24 | Disease | Modulates | Pathway | MONDO | KEGG, Reactome | KEGG, CTD | - | |
| 25 | Disease | Is treated by | Drug | MONDO | Drugbank | ChEMBL, KEGG, CTD | - | |
| 26 | Disease | Caused by | Organism Taxon (pathogen) | MONDO | NCBI Tax ID | PathoPhenoDB | - | |
| 27 | Protein Domain | Enables | Molecular function | InterPro | GO | InterPro2GO | - | |
| 28 | Protein Domain | Localizes to | Cellular component | InterPro | GO | InterPro2GO | - | |
| 29 | Protein Domain | Is involved in | Biological process | InterPro | GO | InterPro2GO | - | |
| 30 | Molecular function/ Cellular component/ Biological process | Is a / Part of / Negatively regulates / Positively regulates | Molecular function/ Cellular component/ Biological process | GO | GO | GOA | - | |
| 31 | Phenotype | Is a | Phenotype | HPO | HPO | HPO | - | |
| 32 | Pathway | Is part of / Is equivalent to / Participates | Pathway | KEGG, Reactome | KEGG, Reactome | Reactome, Compath | - | |
| 33 | Pathway | Is ortholog to | Pathway | KEGG, Reactome | KEGG, Reactome | KEGG, Reactome | Only between human and other organisms | |
| 34 | Side effect | Is a | Side effect | Meddra | Meddra | ADReCS | - | |
| 35 | EC Number | Is a | EC Number | EC Number | EC Number | Expasy ENZYME | - | |
| Source node type | Relationship | Target node type | Source node identifier | Target node identifier | Source database | Filtering (thresholds for filtering) |
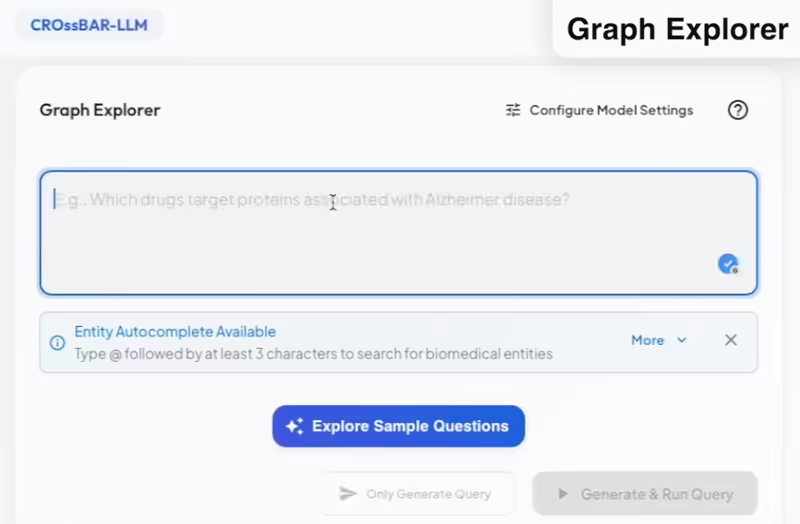
Interact with the CROssBARv2 database using natural language. Through the Graph Explorer, you can navigate direct relationships between entities, retrieving structured facts and connections from the graph. The Semantic Search feature, powered by embeddings, enables the discovery of biologically meaningful patterns by identifying similarities between entities, that go beyond direct graph links.
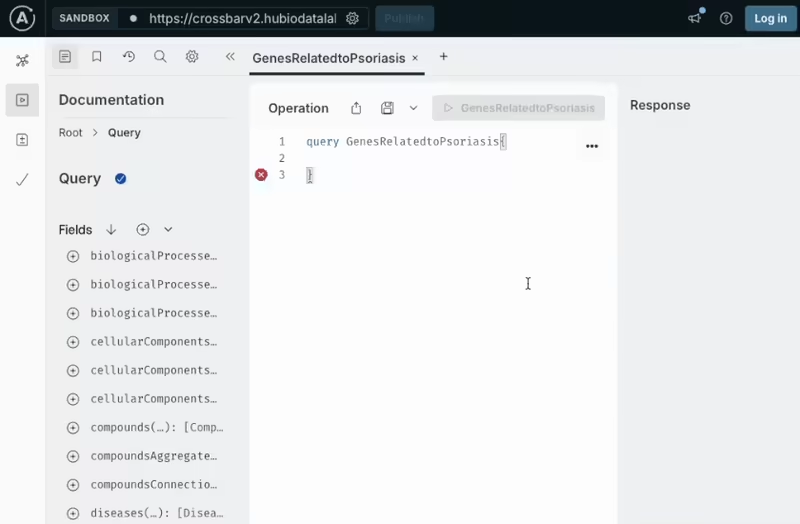
Access the CROssBARv2 database programmatically using a flexible GraphQL interface. You can build custom, nested queries to retrieve precisely the data you need. It's ideal for integrating CROssBARv2 into your analytical workflows or applications, and supports seamless development with tools like Apollo Studio.
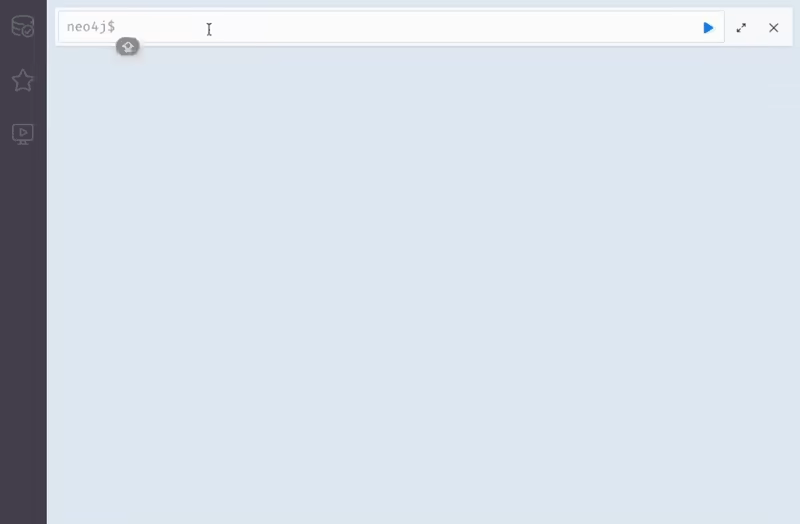
Explore the CROssBARv2 knowledge graph visually through the Neo4j Browser interface. This interactive tool lets you run Cypher queries, visualize nodes and relationships, and investigate the structure of the graph in detail.
1Biological Data Science Lab, Dept. of Computer Engineering, Hacettepe University, Ankara, Turkey
2Dept. of Bioinformatics, Graduate School of Health Sciences, Hacettepe University, Ankara, Turkey
3Dept. of Health Informatics, Institute of Informatics, Hacettepe University, Ankara, Turkey
4Heidelberg University, Faculty of Medicine, and Heidelberg University Hospital, Institute for Computational Biomedicine, Heidelberg, Germany
5Institute of Computational Biology, Computational Health Center, Helmholtz Center, Munich, Germany
6European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Hinxton, Cambridgeshire, UK
* To whom the correspondence should be addressed: Tunca Doğan (tuncadogan@gmail.com)
For further inquiries, please contact us.